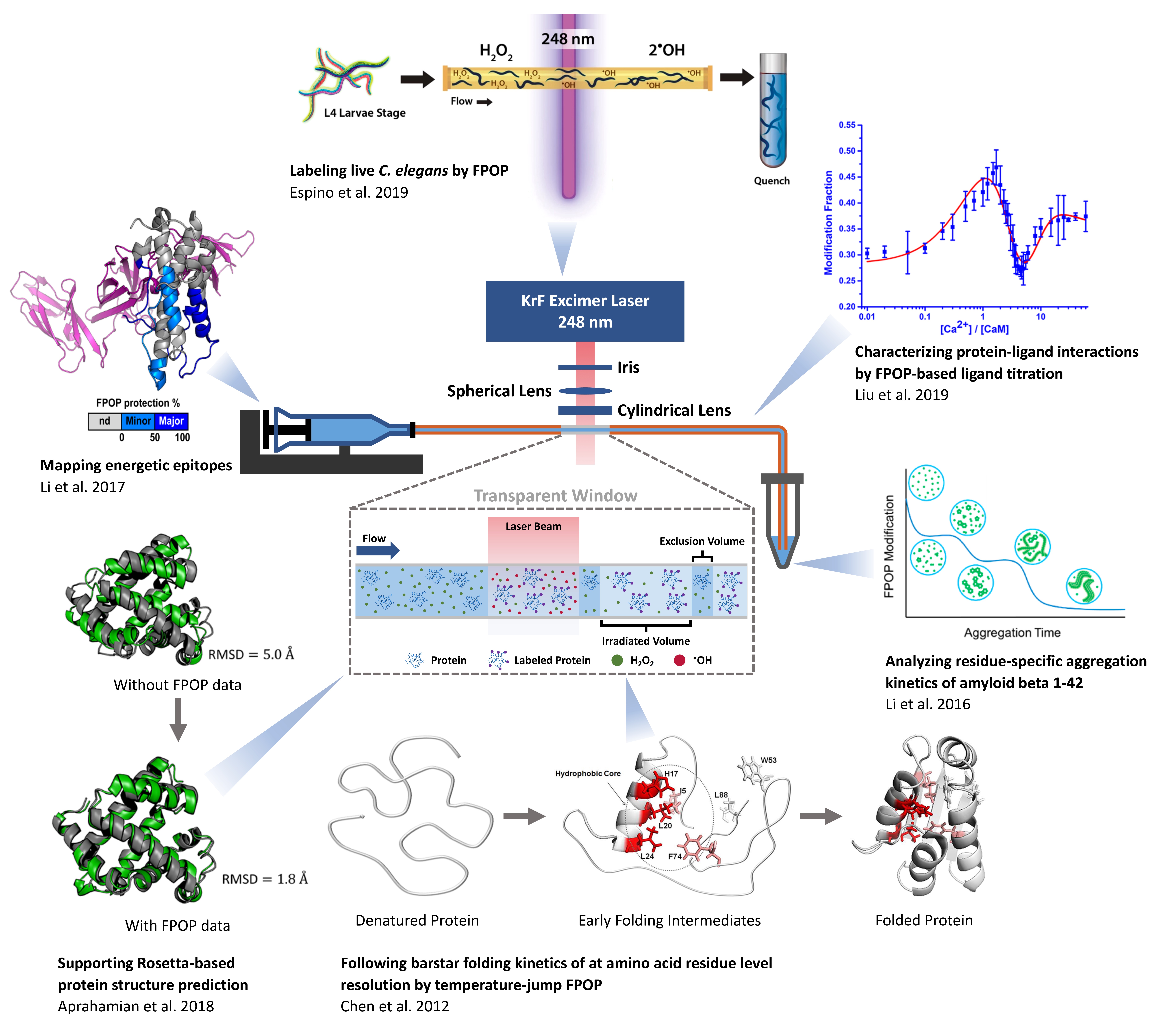
By publishing their method for fast photochemical oxidation of proteins (FPOP), researchers in Michael Gross’s lab have opened doors for fellow scientists to better address research questions related to Alzheimer’s disease, the current coronavirus pandemic, and more.
Michael Gross, professor of chemistry in Arts & Sciences and of immunology and internal medicine at the School of Medicine, and his team are experts in footprinting proteins, using advanced methods for investigating the structure and interactions of proteins within larger molecules. By sharing their method for fast photochemical oxidation of proteins (FPOP), a means of protein footprinting, they hope to support other labs in developing broader applications of FPOP to better address outstanding questions in structural biology. Their work, “Protein higher-order-structure determination by fast photochemical oxidation of proteins and mass spectrometry analysis,” is featured in Nature Protocols and will be freely available for a week starting on Nov 17.
Lead author Roger Liu, a graduate student working with Gross, describes FPOP as a significant advancement in protein footprinting, though it has faced challenges in wider adoption since its development. “FPOP has drawn significant attention because it complements existing footprinting. Its major advantages include fast labeling time frame, irreversible nature, high sensitivity, and relatively broad amino acid residue coverage,” Liu said. Despite the compelling advantages of FPOP, the technical difficulty of establishing the platform has caused a lag in broader applications. Liu cites challenges including choosing the proper laser, setting up the laser optics, establishing the flow system, acquiring the footprint, and analyzing the results by mass spectrometry.
“We always thought that the best way to disseminate FPOP is by applications,” Gross said. “Following its discovery, we have implemented it for problems in biochemistry and biophysics.” Among the applications are following fast protein folding; understanding aggregating proteins with implications in Alzheimer’s disease; mapping epitopes, an important application in our current pandemic; uncovering hidden conformational changes invisible to other structural methods; and determining binding sites and binding affinities of small molecules that bind to proteins. Gross added, “Currently, in a collaboration with Weikai Li in the Department of Biochemistry and Molecular Biophysics, we are moving into transmembrane and membrane-associated proteins where new structural methods are desperately needed for this important class of proteins.”
Publishing their detailed protocol provides other researchers with a recipe for success, hopefully eliminating the technical barrier of adopting FPOP and helping researchers better address research questions in structural biology, biochemistry, and biophysics.
When it comes to the future of FPOP, the team anticipates growth through collaboration and the fresh perspectives it brings. “The FPOP approach will grow as we take ideas from organic chemistry to develop new chemical footprinters for proteins. We hope this protocol that Liu initiated will enable others to adopt the approach. Meanwhile, we welcome collaborators who want us to solve their problem of interest based on our expertise,” they said.



