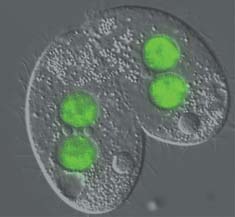
Professor Chalker's laboratory studies a remarkable process of chromosomal rearrangement that occurs during development of the ciliated protozoan Tetrahymena thermophila. His research aims to ultimately learn fundamental principles governing chromosome structure and genetic stability.
The Chalker lab aims to understand the regulation of this massive genome reorganization using a combination of genetic, molecular, and cellular biology approaches to uncover how ~6000 DNA segments are selectively excised. The current model is built on the observations that bi-directional germline transcription leads to the generation of 28-30 base RNA molecules (scan RNAs) that then target specific chromatin modification(s) to the homologous locus. The DNA rearrangement machinery recognizes the modified chromatin state and eliminates the targeted DNA segment.
These studies will certainly provide fundamental insight into RNAi-related mechanisms that direct chromatin modifications that are critical for transcriptional gene silencing and heterochromatin formation in eukaryotes. Underlying this proposal is a goal to understand how RNA molecules can communicate genetic information between the parental and developing genomes, which has great potential to reveal novel roles for RNA in epigenetic programming. Additionally, we believe many of the DNA segments targeted for elimination are important for germline chromosome structure and thus understanding how the cell specifically recognizes these sequences will contribute general knowledge of mechanisms ensuring chromosome stability that are essential to prevent aberrant rearrangements.